Description¶
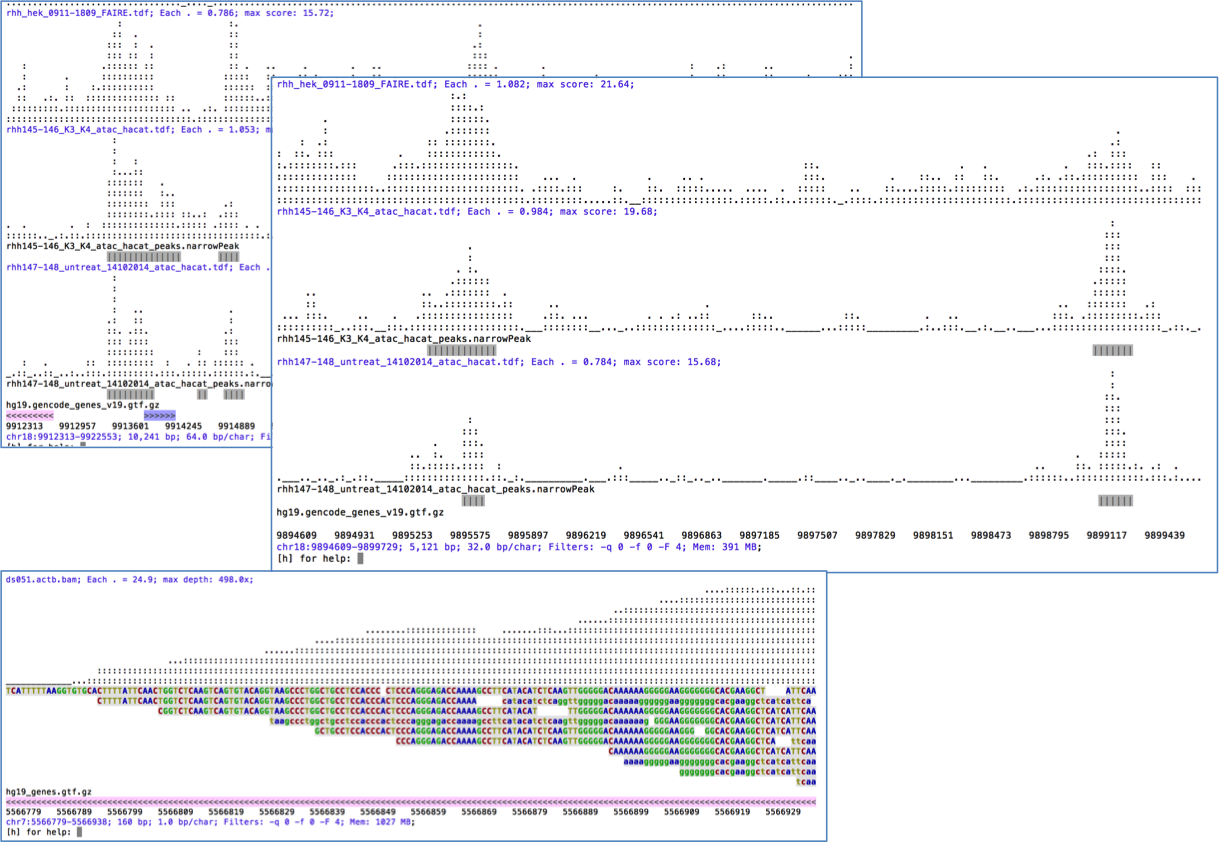
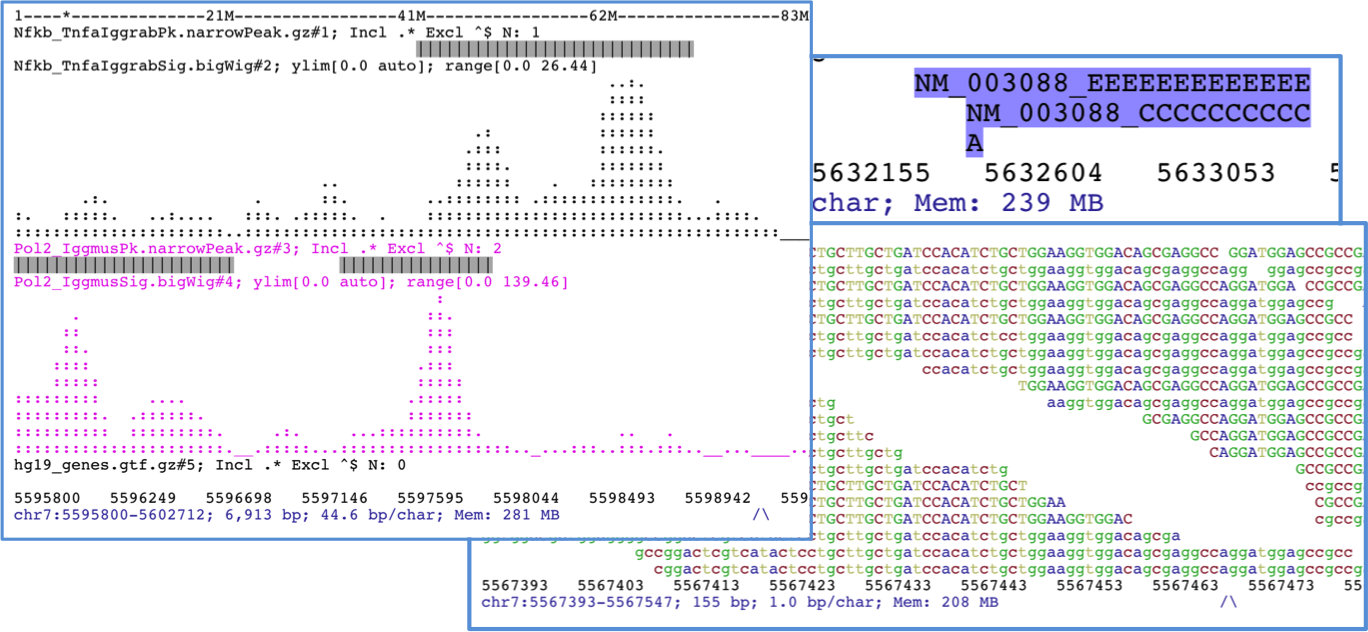
ASCIIGenome is a command-line genome browser running from terminal window and solely based on ASCII characters. Since ASCIIGenome does not require a graphical interface it is particularly useful for quickly visualizing genomic data on remote servers. The idea is to make ASCIIGenome the Vim of genome viewers.
The closest program to ASCIIGenome is probably samtools tview but ASCIIGenome offers much more flexibility, similar to popular GUI viewers like the IGV browser.
Some key features:
- Command line input and interaction, no graphical interface, minimal installation.
- Can load multiple files in various formats.
- Can access remote files via URL or ftp address.
- Easy navigation and searching of features and sequence motifs and filtering options.
- Support for BS-Seq alignment.
Getting help¶
Feel free to send any comment, bug, or issues to one or more of the following:
- Open an issue on GitHub
- Post a question on Biostars.org or bioinformatics.stackexchange.com (recommended: send a notification email to
dario<dot>beraldi<at>gmail<dot>comso I read the question). - Send email to
dario<dot>beraldi<at>gmail<dot>.com
How to cite¶
If you happen to find ASCIIGenome useful and you would like to acknowledge it, please quote the GitHub repository (https://github.com/dariober/ASCIIGenome/).
An article describing ASCIIGenome has been published in Bioinformatics .
Credits¶
- Bam processing is mostly done with the samtools/htsjdk library.
- Bigwig and tdf are processed with classes from IGV source code.
- Block compression and indexing done using jvarkit.
- Brew installation thanks to dalloliogm.